Hepatitis B virus precore mutant
A precore mutant is a variety of hepatitis B virus that does not produce hepatitis B virus e antigen (HBeAg).[1] These mutants are important because infections caused by these viruses are difficult to treat,[2] and can cause infections of prolonged duration and with a higher risk of liver cirrhosis.[3] The mutations are changes in DNA bases from guanine to adenine at base position 1896 (G1896A), and from cytosine to thymine at position 1858 (C1858T) in the precore region of the viral genome.[4]
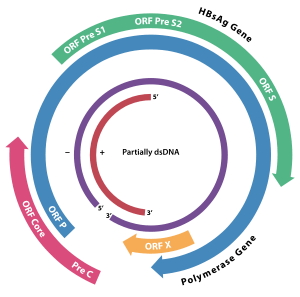
The HBV has four genes: S, P, C, and X. The S gene codes for the "major" envelope protein (HBsAg). The largest gene is P. It codes for DNA polymerase. The C gene codes for HBeAg and HBcAg. The C gene has a precore and a core region. If translation is initiated at the precore region, the protein product is HBeAg. If translation begins with the core region, HBcAg is the protein product. HBeAg is a marker of HBV replication and infectivity. The precore region is not necessary for viral replication. Precore mutants can replicate. They are readily detectable by HBV DNA in serum, but hepatitis B e antigen (HbeAg) is absent. The X gene codes for HBxAg.[5] The product of the X gene is hepatitis B x antigen (HBxAg). It may be involved in carcinogenesis.[6]
Basal core promoter mutants cause a reduction in HBeAg production. These mutations, A1762T and G1764A, occur in the basal core promoter region of the genome.[4] The frequency of these mutations varies according to the HBV genotype.[7]
References
- ↑ Buti M, Rodriguez-Frias F, Jardi R, Esteban R (December 2005). "Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes". Journal of Clinical Virology. 34 Suppl 1: S79–82. doi:10.1016/s1386-6532(05)80015-0. PMID 16461229.
- ↑ Sonneveld MJ, Rijckborst V, Zeuzem S, et al. (July 2012). "Presence of precore and core promoter mutants limits the probability of response to peginterferon in hepatitis B e antigen-positive chronic hepatitis B". Hepatology (Baltimore, Md.). 56 (1): 67–75. doi:10.1002/hep.25636. PMID 22307831.
- ↑ Cleveland Clinic CME hepatitis B Retrieved 15 March 2013
- 1 2 Tacke F, Gehrke C, Luedde T, Heim A, Manns MP, Trautwein C (August 2004). "Basal core promoter and precore mutations in the hepatitis B virus genome enhance replication efficacy of Lamivudine-resistant mutants". Journal of Virology. 78 (16): 8524–35. doi:10.1128/JVI.78.16.8524-8535.2004. PMC 479060
 . PMID 15280461.
. PMID 15280461. - ↑ Karayiannis P, Thomas HC (2009). Mahy BW, van Regenmortel MH, eds. Desk Encyclopedia of Human and Medical Virology. Boston: Academic Press. p. 106. ISBN 0-12-375147-0.
- ↑ Wei Y, Neuveut C, Tiollais P, Buendia MA (August 2010). "Molecular biology of the hepatitis B virus and role of the X gene". Pathologie-biologie. 58 (4): 267–72. doi:10.1016/j.patbio.2010.03.005. PMID 20483545.
- ↑ Lin CL, Kao JH (January 2011). "The clinical implications of hepatitis B virus genotype: Recent advances". Journal of Gastroenterology and Hepatology. 26 Suppl 1: 123–30. doi:10.1111/j.1440-1746.2010.06541.x. PMID 21199523.