Gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage.[1]
Mechanisms of duplication
Ectopic Recombination
Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes.The chance of this happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombination is typically mediated by sequence similarity at the duplicate breakpoints, which form direct repeats. Repetitive genetic elements such as transposable elements offer one source of repetitive DNA that can facilitate recombination, and they are often found at duplication breakpoints in plants and mammals.[2]
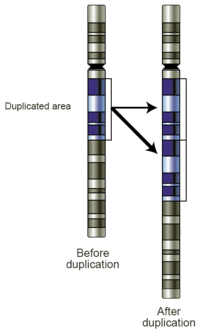
Replication Slippage
Replication slippage is an error in DNA replication that can produce duplications of short genetic sequences. During replication DNA polymerase begins to copy the DNA. At some point during the replication process, the polymerase dissociates from the DNA and replication stalls. When the polymerase reattaches to the DNA strand, it aligns the replicating strand to an incorrect position and incidentally copies the same section more than once. Replication slippage is also often facilitated by repetitive sequences, but requires only a few bases of similarity.
Retrotransposition
During cellular invasion by a replicating retroelement or retrovirus, viral proteins copy their genome by reverse transcribing RNA to DNA. If viral proteins aberrantly attach to cellular mRNA, they can reverse transcribe copies of genes to create retrogenes. Retrogenes usually lack intronic sequences, and often contain poly A sequences that are also integrated into the genome. Many retrogenes display changes in gene regulation in comparison to their parental gene sequences, which sometimes results in novel functions.
Aneuploidy
Aneuploidy occurs when nondisjunction at a single chromosome results in an abnormal number of chromosomes. Aneuploidy is often harmful and in mammals regularly leads to spontaneous abortions (miscarriages). Some aneuploid individuals are viable, for example trisomy 21 in humans, which leads to Down syndrome. Aneuploidy often alters gene dosage in ways that are detrimental to the organism; therefore, it is unlikely to spread through populations.
Whole Genome Duplication
Whole genome duplication, or polyploidy, is a product of nondisjunction during meiosis which results in additional copies of the entire genome. Polyploidy is common in plants, but historically has also occurred in animals, with two rounds of whole genome duplication in the vertebrate lineage leading to humans.[3] After whole genome duplications many sets of additional genes are eventually lost, returning to singleton state. However, retention of many genes, most notably Hox genes, has led to adaptive innovation.
Polyploid is also a well known source of speciation, as offspring, which have different numbers of chromosomes compared to parent species, are often unable to interbreed with non-polyploid organisms. Whole genome duplications are thought to be less detrimental than aneuploidy as the relative dosage of individual genes should be the same.
Gene duplication as an evolutionary event
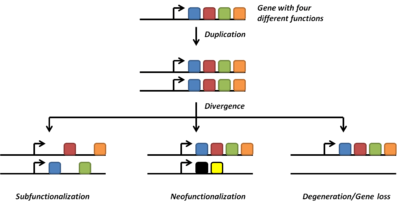
Neofunctionalization
Gene duplications are an essential source of genetic novelty that can lead to evolutionary innovation. Duplication creates genetic redundancy, where the second copy of the gene is often free from selective pressure — that is, mutations of it have no deleterious effects to its host organism. If one copy of a gene experiences a mutation that affects its original function, the second copy can serve as a 'spare part' and continue to function correctly. Thus, duplicate genes accumulate mutations faster than a functional single-copy gene, over generations of organisms, and it is possible for one of the two copies to develop a new and different function. Some examples of such neofunctionalization is the apparent mutation of a duplicated digestive gene in a family of ice fish into an antifreeze gene and duplication leading to a novel snake venom gene [4] and the synthesis of 1 beta-hydroxytestosterone.[5]
Gene duplication is believed to play a major role in evolution; this stance has been held by members of the scientific community for over 100 years.[6] Susumu Ohno was one of the most famous developers of this theory in his classic book Evolution by gene duplication (1970).[7] Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor.[8] Major genome duplication events can be quite common. It is believed that the entire yeast genome underwent duplication about 100 million years ago.[9] Plants are the most prolific genome duplicators. For example, wheat is hexaploid (a kind of polyploid), meaning that it has six copies of its genome.
Subfunctionalization
Another possible fate for duplicate genes is that both copies are equally free to accumulate degenerative mutations, so long as any defects are complemented by the other copy. This leads to a neutral "subfunctionalization" or DDC (duplication-degeneration-complementation) model,[10][11] in which the functionality of the original gene is distributed among the two copies. Neither gene can be lost, as both now perform important non-redundant functions, but ultimately neither is able to achieve novel functionality.
Subfunctionalization can occur through neutral processes in which mutations accumulate with no detrimental or beneficial effects. However, in some cases subfunctionalization can occur with clear adaptive benefits. If an ancestral gene is pleiotropic and performs two functions, often neither one of these two functions can be changed without affecting the other function. In this way, partitioning the ancestral functions into two separate genes can allow for adaptive specialization of subfunctions, thereby providing an adaptive benefit.[12]
Loss
Often the resulting genomic variation leads to gene dosage dependent neurological disorders such as Rett-like syndrome and Pelizaeus-Merzbacher disease.[13] Such detrimental mutations are likely to be lost from the population and will not be preserved or develop novel functions. However, many duplications are, in fact, not detrimental or beneficial, and these neutral sequences may be lost or may spread through the population through random fluctuations via genetic drift.
Identifying duplications in sequenced genomes
Criteria and Single Genome Scans
The two genes that exist after a gene duplication event are called paralogs and usually code for proteins with a similar function and/or structure. By contrast, orthologous genes present in different species which are each originally derived from the same ancestral sequence. (See Homology of sequences in genetics).
It is important (but often difficult) to differentiate between paralogs and orthologs in biological research. Experiments on human gene function can often be carried out on other species if a homolog to a human gene can be found in the genome of that species, but only if the homolog is orthologous. If they are paralogs and resulted from a gene duplication event, their functions are likely to be too different. One or more copies of duplicated genes that constitute a gene family may be affected by insertion of transposable elements that causes significant variation between them in their sequence and finally may become responsible for divergent evolution. This may also render the chances and the rate of gene conversion between the homologs of gene duplicates due to less or no similarity in their sequences.
Paralogs can be identified in single genomes through a sequence comparison of all annotated gene models to one another. Such a comparison can be performed on translated amino acid sequences (e.g. BLASTp, tBLASTx) to identify ancient duplications or on DNA nucleotide sequences (e.g. BLASTn, megablast) to identify more recent duplications. Most studies to identify gene duplications require reciprocal-best-hits or fuzzy reciprocal-best-hits, where each paralog must be the other's single best match in a sequence comparison.[14]
Most gene duplications exist as low copy repeats (LCRs), rather highly repetitive sequences like transposable elements. They are mostly found in pericentronomic, subtelomeric and interstitial regions of a chromosome. Many LCRs, due to their size (>1Kb), similarity, and orientation, are highly susceptible to duplications and deletions.
Genomic microarrays detect duplications
Technologies such as genomic microarrays, also called array comparative genomic hybridization (array CGH), are used to detect chromosomal abnormalities, such as microduplications, in a high throughput fashion from genomic DNA samples. In particular, DNA microarray technology can simultaneously monitor the expression levels of thousands of genes across many treatments or experimental conditions, greatly facilitating the evolutionary studies of gene regulation after gene duplication or speciation.[15][16]
Next generation sequencing
Gene duplications can also be identified through the use of next-generation sequencing platforms. The simplest means to identify duplications in genomic resequencing data is through the use of paired-end sequencing reads. Tandem duplications are indicated by sequencing read pairs which map in abnormal orientations. Through a combination of increased sequence coverage and abnormal mapping orientation, it is possible to identify duplications in genomic sequencing data.
Gene duplication as amplification
Gene duplication does not necessarily constitute a lasting change in a species' genome. In fact, such changes often don't last past the initial host organism. From the perspective of molecular genetics, amplification is one of many ways in which a gene can be overexpressed. Genetic amplification can occur artificially, as with the use of the polymerase chain reaction technique to amplify short strands of DNA in vitro using enzymes, or it can occur naturally, as described above. If it's a natural duplication, it can still take place in a somatic cell, rather than a germline cell (which would be necessary for a lasting evolutionary change).
Role in cancer
Duplications of oncogenes are a common cause of many types of cancer. In such cases the genetic duplication occurs in a somatic cell and affects only the genome of the cancer cells themselves, not the entire organism, much less any subsequent offspring.
| Cancer type | Associated gene amplifications | Prevalence of amplification in cancer type (percent) |
|---|---|---|
| Breast cancer | MYC | 20%[17] |
| ERBB2 (HER2) | 20%[17] | |
| CCND1 (Cyclin D1) | 15–20%[17] | |
| FGFR1 | 12%[17] | |
| FGFR2 | 12%[17] | |
| Cervical cancer | MYC | 25–50%[17] |
| ERBB2 | 20%[17] | |
| Colorectal cancer | HRAS | 30%[17] |
| KRAS | 20%[17] | |
| MYB | 15–20%[17] | |
| Esophageal cancer | MYC | 40%[17] |
| CCND1 | 25%[17] | |
| MDM2 | 13%[17] | |
| Gastric cancer | CCNE (Cyclin E) | 15%[17] |
| KRAS | 10%[17] | |
| MET | 10%[17] | |
| Glioblastoma | ERBB1 (EGFR) | 33–50%[17] |
| CDK4 | 15%[17] | |
| Head and neck cancer | CCND1 | 50%[17] |
| ERBB1 | 10%[17] | |
| MYC | 7–10%[17] | |
| Hepatocellular cancer | CCND1 | 13%[17] |
| Neuroblastoma | MYCN | 20–25%[17] |
| Ovarian cancer | MYC | 20–30%[17] |
| ERBB2 | 15–30%[17] | |
| AKT2 | 12%[17] | |
| Sarcoma | MDM2 | 10–30%[17] |
| CDK4 | 10%[17] | |
| Small cell lung cancer | MYC | 15–20%[17] |
See also
- Pseudogenes
- Molecular evolution
- Unequal crossing over
- Human genome
- Comparative genomics
- Inparanoid
- Tandem exon duplication
References
- ↑ Zhang J (2003). "Evolution by gene duplication: an update". Trends in Ecology & Evolution. 18 (6): 292–8. doi:10.1016/S0169-5347(03)00033-8.
- ↑ "Definition of Gene duplication". medterms medical dictionary. MedicineNet. 2012-03-19.
- ↑ Holland, Peter; Dehal, Paramvir; Boore, Jeffrey L (2005). "Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate". PLoS Biology. 3 (10): e314. doi:10.1371/journal.pbio.0030314. ISSN 1545-7885.
- ↑ Lynch VJ (2007). "Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes". BMC Evolutionary Biology. 7: 2. doi:10.1186/1471-2148-7-2. PMC 1783844
 . PMID 17233905.
. PMID 17233905. - ↑ Conant GC, Wolfe KH (2008). "Turning a hobby into a job: how duplicated genes find new functions". Nature Reviews Genetics. 9 (12): 938–950. doi:10.1038/nrg2482. PMID 19015656.
- ↑ Taylor JS, Raes J (2004). "Duplication and divergence: the evolution of new genes and old ideas". Annu. Rev. Genet. 38: 615–43. doi:10.1146/annurev.genet.38.072902.092831. PMID 15568988.
- ↑ Ohno, S. (1970). Evolution by gene duplication. Springer-Verlag. ISBN 0-04-575015-7.
- ↑ Ohno, S. (1967). Sex Chromosomes and Sex-linked Genes. Springer-Verlag. ISBN 91-554-5776-2.
- ↑ Kellis M, Birren BW, Lander ES (April 2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–24. doi:10.1038/nature02424. PMID 15004568.
- ↑ Force A.; Lynch M.; Pickett F.B.; Amores A.; Yan Y.L.; Postlethwait J. (1999). "Preservation of duplicate genes by complementary, degenerative mutations.". Genetics. 151 (4): 1531–45. PMC 1460548
 . PMID 10101175.
. PMID 10101175. - ↑ Stoltzfus, A. (1999). "On the possibility of constructive neutral evolution". J Mol Evol. 49 (2): 169–181. doi:10.1007/PL00006540. PMID 10441669.
- ↑ Des Marais DL, Rausher MD (2008). "Escape from adaptive conflict after duplication in an anthocyanin pathway gene". Nature. 454 (7205): 762–5. doi:10.1038/nature07092. PMID 18594508.
- ↑ Lee JA, Lupski JR (October 2006). "Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders". Neuron. 52 (1): 103–21. doi:10.1016/j.neuron.2006.09.027. PMID 17015230.
- ↑ Hahn MW, Han MV, Han SG (2007). "Gene Family Evolution across 12 Drosophila Genomes". PLoS Genet. 3 (11): e197. doi:10.1371/journal.pgen.0030197. PMC 2065885
 . PMID 17997610.
. PMID 17997610. - ↑ Mao R, Pevsner J (2005). "The use of genomic microarrays to study chromosomal abnormalities in mental retardation". Ment Retard Dev Disabil Res Rev. 11 (4): 279–85. doi:10.1002/mrdd.20082. PMID 16240409.
- ↑ Gu X, Zhang Z, Huang W (January 2005). "Rapid evolution of expression and regulatory divergences after yeast gene duplication". Proc. Natl. Acad. Sci. U.S.A. 102 (3): 707–12. doi:10.1073/pnas.0409186102. PMC 545572
 . PMID 15647348.
. PMID 15647348. - 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 Kinzler, Kenneth W.; Vogelstein, Bert (2002). The genetic basis of human cancer. McGraw-Hill. p. 116. ISBN 0-07-137050-1.
External links
- A bibliography on gene and genome duplication
- A brief overview of mutation, gene duplication and translocation